MLZ ist eine Kooperation aus:
 > Technische Universität München
> Technische Universität München > Helmholtz-Zentrum Hereon
> Helmholtz-Zentrum Hereon
 > Forschungszentrum Jülich
> Forschungszentrum Jülich
MLZ ist Mitglied in:
 > LENS
> LENS > ERF-AISBL
> ERF-AISBL
MLZ in den sozialen Medien:

MLZ
Lichtenbergstr.1
85748 Garching
Polyethylene glycol polymer layers – studies from tethered lipid bilayers to protein-cell interactions
P. J. F. Röttgermann1, S. Hertrich1, I. Berts1, A. Rühm2, J.-F. Moulin3, J.O. Rädler1, and B. Nickel1
1Fakultät für Physik & CeNS, Ludwig-Maximilians-Universität, München, Germany
2Max Planck Institute for Intelligent Systems, Stuttgart, Germany
3German Engineering Material Science Centre (GEMS) at MLZ, Helmholtz-Zentrum Geesthacht GmbH, Garching, Germany
The structural design and analysis of bio-mimicking surfaces is of great importance for the design of artificial environments for cell adhesion. Such bioavailable surfaces can be mimicked by tethered, solid-supported lipid bilayers (TLB) using polyethylene glycol (PEG) as a cushion. PEG can also be used for the spatial organization of proteins and PEG linked as a copolymer on a surface. Thus, cell motility can be tuned by variation of the surface parameters.
PEG-Tethered Lipid Bilayers
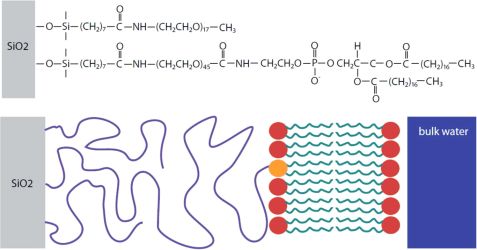
Solid-supported lipid bilayers can act as a workbench for the study of membrane processes. The realization of tethered lipid bilayers is difficult. Here, we designed TLBs with a cushion thickness comparable to a bilayer dimension [1]. LipoPEG was used, as PEG is a weakly interacting cushion material. The PEG end is grafted onto the solid surface and the lipid end can anchor a lipid bilayer (Fig. 1).
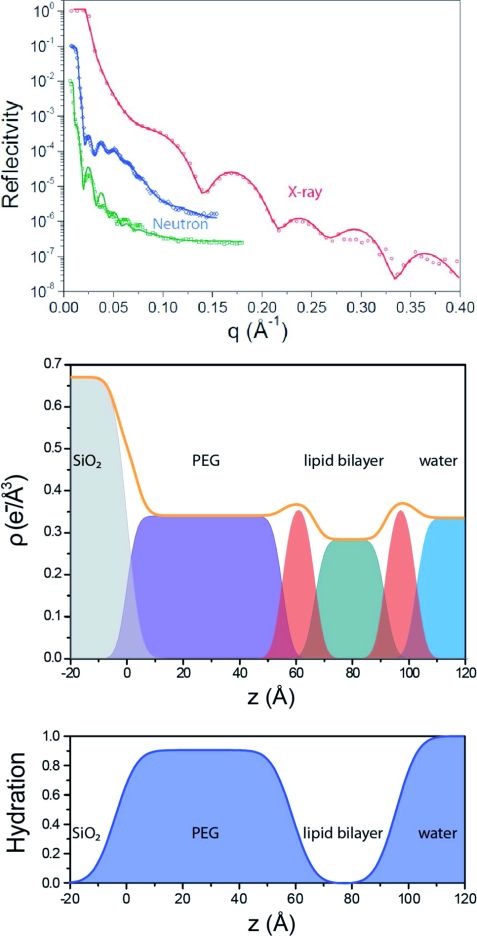
Using FRAP measurements, we determined a diffusion constant of 2.1 ± 0.1 μm2s-1 for the TLB which is only 12 % lower than the diffusion constants of supported lipid bilayers (SLB). This suggests that the bilayer exhibits only a small immobile fraction without any grafted lipids as obstacles. Specular x-ray and neutron reflectivity (NREX at MLZ) were performed to determine the structural layer composition of the TLB. A three layer model consisting of a silicon oxide layer, a PEG layer and a lipid chain region fits the neutron reflectivity (Fig. 2). The PEG interlayer of the TLB is highly hydrated, with a water content of 90 ± 3 %.
AFM indentation measurements at 50 different spots proved (i) the PEG perpendicular layer softness and (ii) its homogeneity along the surface. As the TLB ruptures occur only after deformation of about 40 Å compared to 20 Å for a SLB, the additional deformation can be attributed to the compression of the PEG cushion. Possible applications of such elevated lipid bilayers are, for example, the study of membrane-perforating proteins or binding studies of membrane-associated proteins.
Cell Motility on PEG-Copolymers
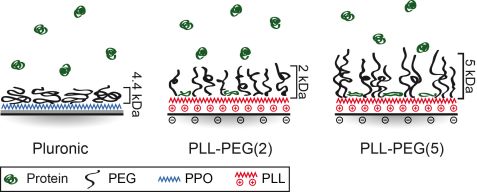
To understand how cells and proteins interact on PEG copolymer surfaces, structural analysis is needed. It has been previously observed that cells are able to migrate on passivated PEG areas which were exposed to proteins such as fibronectin (FN) [2]. Here, we employed neutron scattering (REFSANS at MLZ) to probe the structure of different PEG layers and determine the amount and distribution of FN in these PEG layers [3]. We focused our study on two polymer constructs: first, PEG (4.4 kDa) grafted onto a hydrophobic polymer anchor polypropylene oxide (PPO), and second, PEG chains of 2 and 5 kDa grafted in a ratio of 3.5 onto the poly-L-lysine (PLL-PEG and PLL-PEG). The REFSANS data revealed that PEG repellence is mainly influenced by the underlying polymer-layer and secondly by the PEG density. Hydrophobic layers such as PPO are shielded by the PEG chains and are, therefore, highly protein repellent. In the moiety of hydrophilic layers (PLL) protein adsorption of 0.4 mg/cm² for PLL-PEG and 0.7 mg/cm² for PLL-PEG was found. Higher PEG densities lead to higher protein repellence. However, too high a PEG density (e.g. surface density σ = 1.99 Å-2 for PLL-PEG) can lead to a contrary effect of increased protein adsorption. PEG brushes probably become stiffer and are not that flexible anymore (Fig. 3).
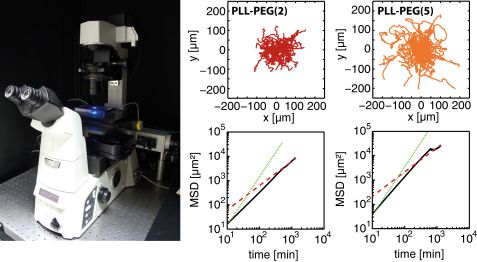
Surface properties were compared with cell behavior. No cell adhesion was noticed as no protein is adsorbed on the PEG linked PPO polymer (Pluronic). By contrast, cell spreading was observed on PLL-PEG. Cell spreading on PLL-PEG is comparable to that on pure FN surfaces. Apart from different spreading, cell motility also correlates with different surface properties. Cell motion was tracked by time-lapse fluorescence microscopy (Fig. 4). The longest persistence time of cell motion is observed on a pure FN surface, whereas the highest velocity is observed on PLL PEG (Fig. 4). As cells polarize in one direction (mostly random in the absence of a chemo-attractant), they have a higher probability of moving forward in the same direction instead of turning towards any other direction if the amount of protein is high. The highest speed is measured on PLL PEG. It exhibits fewer adhesion points than pure FN layers which leads on the one hand to more frequent interruption in the migration and on the other hand to faster movement, possibly due to faster detachment.
We generated artificial surfaces with TLBs which could provide a more natural and cell- like surrounding. Further studies on the membrane interaction of living cells on solid surfaces containing FN and PEG are needed, using neutron reflectivity. Information on the surface interaction of macromolecular biomolecules paves the way for more advanced micro-structured surfaces which can be used for cell migration assays or high-throughput single cell analysis.
Financial support by the Deutsche Forschungs-gemeinschaft (DFG) via project B1 within the SFB 1032, the Excellence Cluster ‘Nanosystems Initiative Munich (NIM)’, the Center for NanoScience (CeNS), FP7 EU grants NanoTransKinetics and NanoMILE, and by BMBF-05K13WM1 and 05K10WM1 is gratefully acknowledged.
References:
[1] S. Hertrich et al.; Langmuir 30, 9442 (2014).
[2] P. J. Röttgermann et al.; Soft Matter 10, 2397 (2014).
[3] P. J. Röttgermann et al.; Macromol. Biosci. 14, 1755 (2014).
MLZ ist eine Kooperation aus:
 > Technische Universität München
> Technische Universität München > Helmholtz-Zentrum Hereon
> Helmholtz-Zentrum Hereon
 > Forschungszentrum Jülich
> Forschungszentrum Jülich
MLZ ist Mitglied in:
 > LENS
> LENS > ERF-AISBL
> ERF-AISBL
MLZ in den sozialen Medien: