MLZ ist eine Kooperation aus:
 > Technische Universität München
> Technische Universität München > Helmholtz-Zentrum Hereon
> Helmholtz-Zentrum Hereon
 > Forschungszentrum Jülich
> Forschungszentrum Jülich
MLZ ist Mitglied in:
 > LENS
> LENS > ERF-AISBL
> ERF-AISBL
MLZ in den sozialen Medien:

MLZ
Lichtenbergstr.1
85748 Garching
Studying the interactions between liposomes and silica nanoparticles using SANS, NSE and DLS
I. Hoffmann1,2, R. Michel1, M. Sharp2, O. Holderer3, M.-S. Appavou3, F. Polzer4, B. Farago2, and M. Gradzielski1
1Stranski-Laboratorium für Physikalische und Theoretische Chemie, Institut für Chemie, Technische Universität Berlin, Berlin, Germany
2Institut Max von Laue-Paul Langevin (ILL), Grenoble, France
3Jülich Centre for Neutron Science (JCNS) at MLZ, Forschungszentrum Jülich GmbH, Garching, Germany
4TEM Group, Institute of Physics, Humboldt Universität zu Berlin, Berlin, Germany
We studied the interactions between DOPC liposomes and small silica nanoparticles (SiNPs). To do so, we per- formed SANS, NSE, DLS and cryo-TEM measurements. The interactions between liposomes and nanoparticles are of fundamental scientific interest as liposomes can serve as model systems for biological cells. Therefore, these measurements are important for an understanding of the effect of nanoparticles on living cells. Using SANS, it could be shown that the SiNPs have almost no effect on the structure of the liposomes despite almost complete adsorption, as evidenced by DLS and TEM. We proceeded to investigate the influence of the SiNPs on the liposomes’ membrane dynamics using NSE and found that the membrane is softened by the binding of the SiNPs to the membrane surface. This surprising result may help in future understanding of the effects of nanoparticles on biological cells.
Vesicles are closed bilayers, dividing an interior part from the bulk solution, very much like cells. If the bilayer consists of phospholipids, as does most of any biological membrane, they are referred to as liposomes and serve as simple model systems for biological cells [1].
With the increasingly wide-spread use of nanoparticles in our daily environment, their interactions with cells have gained quite some interest, whether it is to be able to assess their risks (nanotoxicity) or their benefits in nano medicine [2-4].
To fully understand the effect of nanoparticles on the cell membrane, it is not only necessary to investigate the influence on the structure but also on the dynamics and the stiffness of the membrane.
We have investigated the interactions between liposomes consisting of the phospholipid DOPC (1,2-Dioleoyl-sn-glycero-3-phosphocholine, 0.1 wt%), prepared by extrusion through a 100 nm membrane and SiNPs at a concentration of 0.085 wt% with a radius of 8.4 nm in dilute aqueous solution. To do so, several complementary methods were employed [5].
Structure of the Liposomes
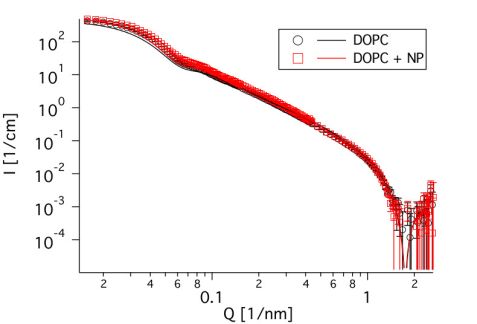
To study the influence on the membrane dynamics, it was necessary to investigate an interacting system, which does not undergo a major structural reorganization. As the contrast of silica is relatively low in neutron scattering, small angle neutron scattering (SANS) is well suited to examination of the structure of the liposomes. Therefore, measurements were performed on the instrument KWS 2 at MLZ to verify the size of the liposomes and their structural stability against the SiNPs. As can be seen in Fig. 1, the curves look almost identical, regardless of the presence or absence of the nanoparticles, which means that the structure of the liposomes is not altered. Furthermore, the size of the liposomes can be determined to be 43.5 nm, as can be seen from the kink at about 0.07 1/nm. This confirms results obtained from cryo-TEM. The identical position of the dip at about 2 1/nm shows that the thickness of the membrane was not changed, either.
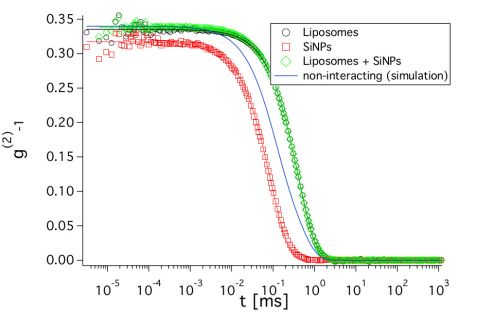
From the SANS measurements alone, it is not clear if the SiNPs are adsorbed on the liposomes. Therefore, dynamic light scattering (DLS) measurements were performed (Fig. 2). They allow one to determine the translational diffusion coefficients of objects in solution, which are related to the sizes of the diffusing objects. The advantage of knowing the translational diffusion coefficient is two-fold. First, it makes it possible to check if the SiNPs are adsorbed and second, translational diffusion also contributes to the signal measured in NSE. Knowing it a priori makes the determination of the membranes` bending rigidity from NSE more reliable. As the SiNPs have a relatively high contrast in light scattering, the fast diffusion of unbound SiNPs would lead to a significantly faster decay of the curve for the sample with liposomes and SiNPs. The simulated curve in Fig. 2 corresponds to the combination of the individual curves from liposomes and SiNPs weighted with their respective intensities, which would result if the SiNPs were not adsorbed on the liposomes. However, the measured curves are almost identical for the liposomes with and without added SiNPs. This indicates that they are bound to the vesicles and further confirms our finding from SANS that the structure of the liposomes is not changed. The diffusion coefficient determined from the decay of the DLS curves will be used in the analysis of the NSE measurements.
Membrane Dynamics
NSE allows the dynamics of objects on a timescale from less than a nanosecond to several hundred nanoseconds and on a size range from less than a nanometer to tens of nanometers to be monitored. It is the neutron scattering method that allows the smallest changes in energy (longest time scales) to be measured. Its size and energy range are well suited to an investigation of the dynamics of biological membranes. Therefore, we performed NSE measurements on the instruments J-NSE (MLZ) and IN15 (ILL).
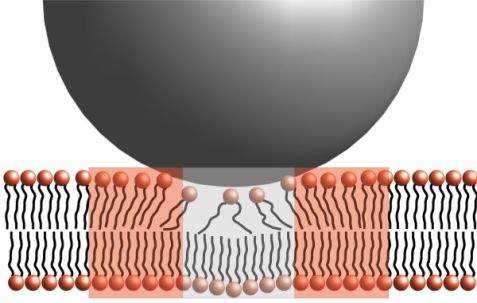
The data could be described as a combination of translational diffusion and membrane undulations in the framework of the Zilman-Granek model6, which predicts the bending modulus κ to scale with the experimentally determined relaxation rate Γ as Γ~(1/κ)0.5. Using the translational diffusion coefficient previously determined by DLS, the only free parameter is the bending rigidity and it was found that the bending rigidity decreases upon addition of the SiNPs. This is a surprising result, as the adsorption of rigid particles would be expected to stiffen the membrane. However, the adsorption of the SiNPs introduces disorder in the bilayer, which softens it in the vicinity of the adsorption site as depicted in Fig. 3. Together with the stiffening of the membrane directly at the adsorption site, this leads to a small but pronounced net softening of the membrane.
References:
[1] D. D. Lasic, Liposomes: from physics to applications, Elsevier Amsterdam (1993).
[2] R. Michel and M. Gradzielski, Int. J. Mol. Sci. 13, 11610 (2012).
[3] R. Michel et al, Soft Matter 9, 4167 (2013).
[4] A. H. Bahrami et al., Adv. Colloid Interface Sci. 208, 214 (2014).
[5] I. Hoffmann et al., Nanoscale 6, 6945 (2014).
[6] A. G. Zilman and R. Granek, Phys. Rev. Lett. 77, 4788 (1996).
MLZ ist eine Kooperation aus:
 > Technische Universität München
> Technische Universität München > Helmholtz-Zentrum Hereon
> Helmholtz-Zentrum Hereon
 > Forschungszentrum Jülich
> Forschungszentrum Jülich
MLZ ist Mitglied in:
 > LENS
> LENS > ERF-AISBL
> ERF-AISBL
MLZ in den sozialen Medien: