MLZ ist eine Kooperation aus:
 > Technische Universität München
> Technische Universität München > Helmholtz-Zentrum Hereon
> Helmholtz-Zentrum Hereon
 > Forschungszentrum Jülich
> Forschungszentrum Jülich
MLZ ist Mitglied in:
 > LENS
> LENS > ERF-AISBL
> ERF-AISBL
MLZ in den sozialen Medien:

MLZ
Lichtenbergstr.1
85748 Garching
Internal nanosecond dynamics in the intrinsically disordered myelin basic protein
A. Stadler1, L. Stingaciu2, A. Radulescu3, O. Holderer3, M. Monkenbusch1, R. Biehl1, and D. Richter1
1Jülich Centre for Neutron Science (JCNS) and Institute for Complex Systems (ICS), Forschungszentrum Jülich GmbH, Jülich, Germany
2Jülich Centre for Neutron Science (JCNS) at SNS, Forschungszentrum Jülich GmbH, Oak Ridge, USA
3Jülich Centre for Neutron Science (JCNS) at MLZ, Forschungszentrum Jülich GmbH, Garching, Germany
Intrinsically disordered proteins lack a well-defined folded structure and contain a high degree of structural freedom and conformational flexibility. In solution, the myelin basic protein belongs to that class of proteins. Using small-angle scattering, the protein was found to be structurally disordered, similar to ideal Gaussian chains. Modelling via a coarse-grained structural ensemble indicated a compact core with flexible ends. Neutron spin-echo spectroscopy measurements revealed a large contribution of internal dynamics to the overall diffusion. In an alternative approach, we investigated whether models from polymer theory are suitable for the interpretation of the observed motions.
Intrinsically Disordered Proteins
The expected structural and dynamic properties of intrinsically disordered proteins (IDPs) range from very soft structures, through folded elements connected by extended and flexible loops, to fully disordered polypeptide chains. Crystallographic structures of IDPs do not exist due to the existence of a large number of different conformational states. However, at low resolution the protein structure in solution can be well characterized by small-angle scattering of X-rays (SAXS) or neutrons (SANS), while neutron spin-echo spectroscopy (NSE) is a method well-suited to the study of polymer dynamics [1] and functional relevant motions of protein domains [2].
The myelin basic protein (MBP) is a major component of the myelin sheath in the central nervous system. In aqueous solution, MBP is primarily unstructured and is classified as intrinsically disordered. In this context, we investigated the nature and extent of large conformational motions in MBP as an example of their role in IDPs [3].
Small-Angle Scattering and NSE Experiments
SANS was measured on the instrument KWS-1 at the MLZ in Garching. SAXS was measured on the instrument BM29 at the ESRF, Grenoble, France. NSE measurements were carried out at the J-NSE spectrometer at the MLZ.
Solution Structure of MBP
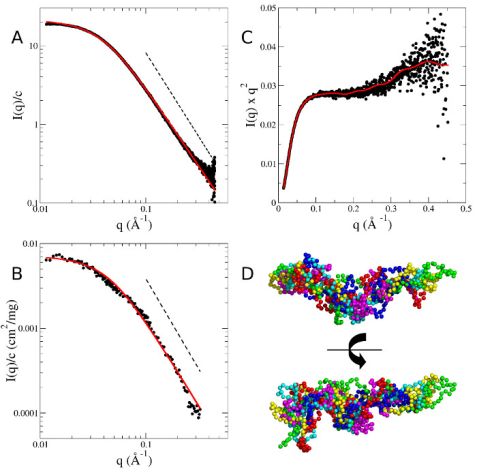
Small-angle scattering was measured to gain information about the solution structure of MBP as a prerequisite for the NSE experiments. Measured SAXS and SANS form factors of MBP are given in Fig. 1. Both curves show power law scattering at q > 0.1 Å-1 with a power law coefficient of -2.1. A power law coefficient of -2 is the characteristic sign of Gaussian chain polymers in Θ solvent and in the melt. The slightly steeper slope of the measured data indicates a more compact conformation as compared to a Gaussian chain. The SAXS and SANS curves measured can be described using the Debye equation for Gaussian chains. Reverse Monte Carlo simulations were applied to generate a coarse-grained ensemble representing the structural characteristics of MBP. The general features of the selected coarse-grained conformations indicate a central core region with flexible termini, while the overall shape appears to be slightly bent (Fig. 1 (D)).
Dynamics of MBP Measured by NSE
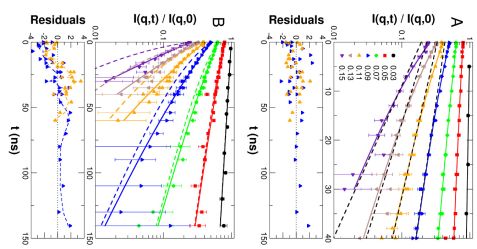
Collective motions of MBP were explored using NSE. The measured NSE spectra of a 54 mg/mL solution are shown in Fig. 2. In general, protein dynamics measured by NSE in solution consist of global diffusion and internal conformational motions. In a first approach we use the structural ensemble determined by small-angle scattering to interpret the NSE spectra, see Fig. 2 (A). Typically, the observed relaxation due to internal protein dynamics decays significantly faster than global protein diffusion, which is the remaining contribution to the NSE spectra in the long-time limit. At shorter times the contribution of internal protein dynamics to the spectra becomes directly visible as compared to the long time limit.
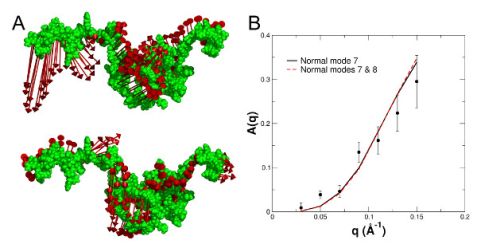
To interpret the observed internal protein dynamics in more detail, the lowest lying collective excitations of the structural ensemble were calculated using normal mode analysis. The calculated displacement patterns of the first and second non-trivial normal modes (NMs 7 and 8) of a representative conformation are shown in Fig. 3 (A). The normal modes were used to describe the measured contribution of internal dynamics A(q) to the NSE spectra, see Fig. 3 (B). NM 7 is the dominating collective excitation of the protein and fully reproduces the measured A(q).
In an alternative approach we tested whether the dynamics of MBP can be described using simplified models from polymer theory. The classical Zimm model is a coarse-grained description of the dynamics of polymers in solution. To account for additional microscopic interactions – such as, for example, internal barriers, hindered dihedral rotations, side-chain interactions, or hydrogen bonding – the Zimm model was extended to the Zimm models with internal friction (ZIF). The fits with the Zimm and ZIF models to the NSE spectra are shown in Fig 2 (B). The classical Zimm model does not represent the measured data. The ZIF model shows small but systematic deviations from the measured data. The large value of the internal friction also leads to the breakdown of the mathematical structure of the Zimm model.
Conclusions
The NSE experiments showed a high flexibility of the structural ensemble. Our results are important for a biophysical understanding of the nature and extent of large-scale conformational motions in IDPs. Our experiment clearly demonstrates the potential of neutron scattering for the investigation of IDPs.
References:
[1] D. Richter et al., Advances in Polymer Science: J. Neutron Spin Echo in Polymer Systems, Vol. 174, Springer, Berlin (2005).
[2] R. Inoue et al., Biophys. J. 99, 2309 (2010).
[3] A. M. Stadler et al., J. Am. Chem. Soc. 136(19), 6987 (2014).
MLZ ist eine Kooperation aus:
 > Technische Universität München
> Technische Universität München > Helmholtz-Zentrum Hereon
> Helmholtz-Zentrum Hereon
 > Forschungszentrum Jülich
> Forschungszentrum Jülich
MLZ ist Mitglied in:
 > LENS
> LENS > ERF-AISBL
> ERF-AISBL
MLZ in den sozialen Medien: