MLZ is a cooperation between:
 > Technische Universität München
> Technische Universität München > Helmholtz-Zentrum Hereon
> Helmholtz-Zentrum Hereon
 > Forschungszentrum Jülich
> Forschungszentrum Jülich
MLZ is a member of:
 > LENS
> LENS > ERF-AISBL
> ERF-AISBL
MLZ on social media:

MLZ (eng)
Lichtenbergstr.1
85748 Garching
BIODIFF
Diffractometer for large unit cells

This instrument is focussed on cold neutrons. Therefore, please carefully check the “Technical data WITHOUT cold source” section. Deviating parameters are in bold. The instrument team is happy to answer any further questions!
BIODIFF is designed to handle crystals with large unit cells and is dedicated to the structure determination of biological macromolecules.
In biological macromolecules, like proteins and nucleic acids, hydrogen atoms play an important role. Hydrogen atoms participate in the substrate binding process and are essential for proton transfer reactions during the catalysis in many enzymes. Therefore, the knowledge about the protonation states of amino acid residues in the active centre of proteins is often crucial for the understanding of their reaction mechanisms. However, hydrogen atoms, especially rather flexible ones, are barely detectable in X-ray structure determinations of proteins. On the other hand, hydrogen atoms are clearly visible in neutron crystallography experiments even at moderate resolutions (dmin < 2.5 Å).
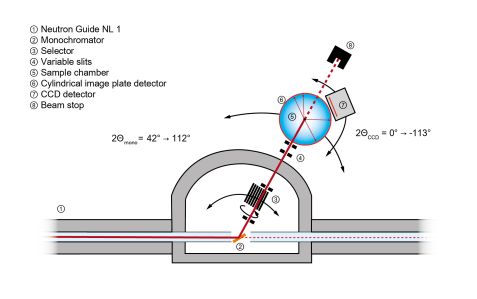
BIODIFF is the first instrument along the neutron guide NL1. Using a pyrolytic graphite monochromator, the diffractometer covers a tunable wavelength range of 1.85 to 5.3 Å. Higher-order wavelength contaminations are removed by a neutron velocity selector. Using the PG(002) reflection, a wavelength range between 2.7 and 5.3 Å can be realised. The wavelength range between 1.85 and 2.7 Å is covered by the PG(004) reflection.
The main detector of the diffractometer consists of a neutron imaging plate system in a cylindrical geometry to cover a large solid angle. A fast LiF/ZnS scintillator CCD camera is foreseen for additional detection abilities.
The main advantage of this instrument is the possibility of adapting the wavelength to the size of the sample crystal’s unit cell while operating with a clean, monochromatic beam that keeps the background level low.
The main field of application is the neutron structure analysis of proteins, especially the determination of hydrogen atom positions.
Typical questions in this field of interest are:- Enzymatic mechanism (protonation states of amino acids)
- Ligand binding mediated by hydrogen bonds
- Investigation of the hydration shell of proteins
- H/D-exchange pattern as a monitor of structural stability/ accessibility/ flexibility
Using a wavelength of 4.7 Å, unit cells with lattice constants up to 200 Å can be measured.
- Oxford Cryosystems Cryostream 700 plus: 90 – 500 K
- Closed cycle cryostat: 3.5 – 325 K
- Neutron guide NL1; supermirror m = 2
- Monochromator:
- Pyrolytic graphite (PG), mosaicity: 0.4 – 0.5°
- Higher order filter:
- Astrium type velocity selector
- Transmission 87 % for 2.7 Å
- Beam characteristic for a horizontal and vertical divergence of 0.5° FWHM:
- 1.85 Å with PG(004): 2.6 × 106 n cm-2 s-1, ∆λ/λ = 1.1 %
- 2.7 Å with PG(002): 2.7 × 106 n cm-2 s-1, ∆λ/λ = 2.1 %
- 3.4 Å with PG(002): 1.3 × 106 n cm-2 s-1, ∆λ/λ = 1.6 %
- 4.7 Å with PG(002): 2.4 × 105 n cm-2 s-1, ∆λ/λ = 1.1 %
- Collimation by adjustable slits between 1 – 4 mm
- Beam divergence (no slits):
- 0.8° FWHM horizontal, 0.7° FWHM vertical
- BaFBr:Eu2+ mixed with Gd2O3
- Dimensions:
- Radius: 200 mm
- Angular range:
- ±152° horizontal
- ±48° vertical
- Pixel size (quadratic): 125, 250, 500 µm
- Readout time (with erasing): 7 min (for 250 µm pixel size)
- ZnS mixed with 6LiF
- Dimensions:
- Active scintillator area (flat): 200 × 200 mm²
- Distance to sample: 100 mm
- 2Θ-angle around sample position 0° – 113°
- CCD chip with 2048 × 2048 pixels
- Pixel size: 13.5 × 13.5 µm2
- Overall spatial resolution: ≈ 300 × 300 µm2 (limited by scintillator thickness)
- Minimum readout time: ≈ 1 sec (full resolution); < 1 sec (binning mode)
- Neutron guide NL1; supermirror m = 2
- Monochromator:
- Pyrolytic graphite (PG), mosaicity: 0.4 – 0.5°
- Higher order filter:
- Astrium type velocity selector
- Transmission 87 % for 2.7 Å
- Beam characteristic for a horizontal and vertical divergence of 0.5° FWHM:
- 1.85 Å with PG(004): 2.2 × 106 n cm-2 s-1, ∆λ/λ = 1.1 %
- 2.7 Å with PG(002): 6.4 × 106 n cm-2 s-1, ∆λ/λ = 2.1 %
- 3.4 Å with PG(002): 5.8 × 106 n cm-2 s-1, ∆λ/λ = 1.6 %
- 4.7 Å with PG(002): 3.3 × 106 n cm-2 s-1, ∆λ/λ = 1.1 %
- Collimation by adjustable slits between 1 – 4 mm
- Beam divergence (no slits):
- 0.8° FWHM horizontal, 0.7° FWHM vertical
- BaFBr:Eu2+ mixed with Gd2O3
- Dimensions:
- Radius: 200 mm
- Angular range:
- ±152° horizontal
- ±48° vertical
- Pixel size (quadratic): 125, 250, 500 µm
- Readout time (with erasing): 7 min (for 250 µm pixel size)
- ZnS mixed with 6LiF
- Dimensions:
- Active scintillator area (flat): 200 × 200 mm²
- Distance to sample: 100 mm
- 2Θ-angle around sample position 0° – 113°
- CCD chip with 2048 × 2048 pixels
- Pixel size: 13.5 × 13.5 µm2
- Overall spatial resolution: ≈ 300 × 300 µm2 (limited by scintillator thickness)
- Minimum readout time: ≈ 1 sec (full resolution); < 1 sec (binning mode)
Instrument scientists
Dr. Andreas Ostermann
Phone: +49 (0)89 289-14702
E-mail: Andreas.Ostermann@frm2.tum.de
Dr. Tobias Schrader
Phone: +49 (0)89 158860-743
E-mail: t.schrader@fz-juelich.de
BIODIFF
Phone: +49 (0)89 289-14565
Operated by


Funding


Publications
Find the latest publications regarding BIODIFF in our publication database iMPULSE:
Citation templates for users
In all publications based on experiments on this instrument, you must provide some acknowledgements. To make your work easier, we have prepared all the necessary templates for you on this page.
Instrument control
Gallery









MLZ is a cooperation between:
 > Technische Universität München
> Technische Universität München > Helmholtz-Zentrum Hereon
> Helmholtz-Zentrum Hereon
 > Forschungszentrum Jülich
> Forschungszentrum Jülich
MLZ is a member of:
 > LENS
> LENS > ERF-AISBL
> ERF-AISBL
MLZ on social media: