MLZ is a cooperation between:
 > Technische Universität München
> Technische Universität München > Helmholtz-Zentrum Hereon
> Helmholtz-Zentrum Hereon
 > Forschungszentrum Jülich
> Forschungszentrum Jülich
MLZ is a member of:
 > LENS
> LENS > ERF-AISBL
> ERF-AISBL
MLZ on social media:

MLZ (eng)
Lichtenbergstr.1
85748 Garching
Neutron cryo-crystallography sheds light on heme peroxidases reaction pathway
C. Casadei1,2, M. Blakeley2, T. Schrader3, A. Ostermann4, E. Raven5, and P. Moody1
1Department of Biochemistry and Henry Wellcome Laboratories for Structural Biology, University of Leicester, Leicester, UK
2Institut Laue-Langevin (ILL), Grenoble, France
3Jülich Centre for Neutron Science (JCNS) at MLZ, Forschungszentrum Jülich GmbH, Garching, Germany
4Heinz Maier-Leibnitz Zentrum (MLZ), Technische Universität München, Garching, Germany
5Department of Chemistry, University of Leicester, Leicester, UK
Heme peroxidases are a family of catalytic iron-containing proteins that are found in nearly all living organisms. These enzymes catalyze the H2O2-dependent oxidation of a substrate, thereby removing this potentially hazardous molecule from the cell. Heme peroxidases share a common reaction mechanism that involves the presence of two intermediate species known as Compound I and Compound II. Compound I contains an oxidized ferryl heme, plus either a porphyrin π-radical or a protein radical. Reduction of Compound I by one electron equivalent yields the closely related Compound II intermediate. Heme peroxidases have been the object of extensive studies in the last decades: of particular interest is the structural characterization of the active site in the transient Compound I. The protonation state of the iron bound oxygen ligand in Compound I has become a key question in the study of heme enzymes, due to its implications for the reaction mechanism. In particular, attention has been focused on whether the ferryl can be formulated as Fe(IV)=O or Fe(IV)-OH.
Neutron cryo-crystallography – the method of choice
The methodologies that were traditionally employed to address this question appeared to be inadequate. Early approaches to the problem used resonance Raman methods to examine the iron-oxygen bond as an indirect reporter on the oxygen protonation state. However, the photolability of Compound I during laser excitation is well documented and results in ambiguous experimental findings. More recently, X-ray crystallography was employed with the purpose of inferring the ligand protonation state from the study of the iron-oxygen distance. However, the catalytic centre in these proteins is particularly sensitive to radiation damage effects and X-ray determined Fe-O distances are now considered unreliable.
For these reasons we adopted a different approach. Neutron crystallography allows the localization of deuterium substituted hydrogen atoms in medium resolution nuclear scattering density maps. By contrast, hydrogen atoms are localized in X-ray maps only at high resolutions of 1.2 Å or beyond. The high X-ray dose required for ultra-high resolution data collection cannot be employed in the study of heme enzymes due to their sensitivity to radiation damage.
Due to the lack of radiation damage effects and its capability of localizing deuterons, neutron crystallography is an excellent tool for the study of hydrogen related biochemistry such as the determination of the protonation state of key residues and ligands, the position of water molecules in the active site and the study of hydrogen bond networks. Neutron crystallography data collection from cryo-trapped reaction intermediates is a unique tool for probing reaction mechanisms, but presents a number of challenges, in particular the need to flash cool the large crystals required for neutron crystallography down to the cryogenic temperature required for the study of transient species.
Structure determination of Compound I
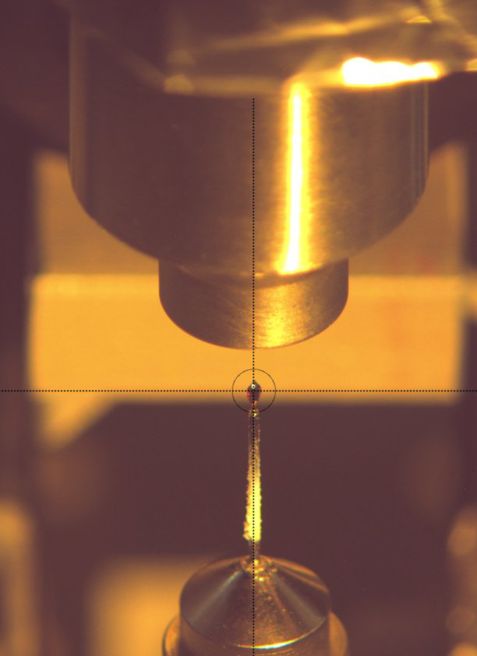
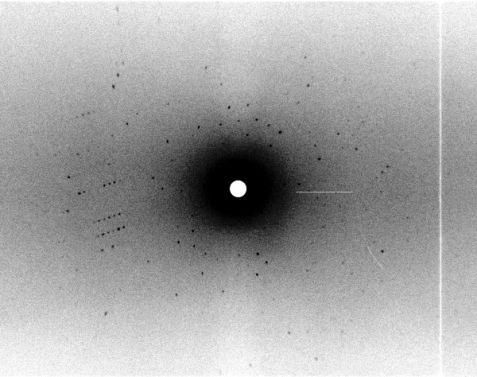
We determined the neutron structure of the transient Compound I of the heme enzyme Cytochrome c Peroxidase (CcP) at 100 K: a deuterium exchanged CcP single crystal was reacted to form Compound I and subsequently cryo-cooled at 100 K (see Fig. 1). Monochromatic neutron data were collected at the instrument BIODIFF at the MLZ (see Fig. 2). We obtained the first cryo-trapped enzyme intermediate structure determined by neutron crystallography. For a direct comparison we also determined the neutron structure of CcP in the resting state, using the quasi-Laue diffractometer LADI III, at the ILL. The data were collected at room temperature on a deuterium exchanged single crystal.
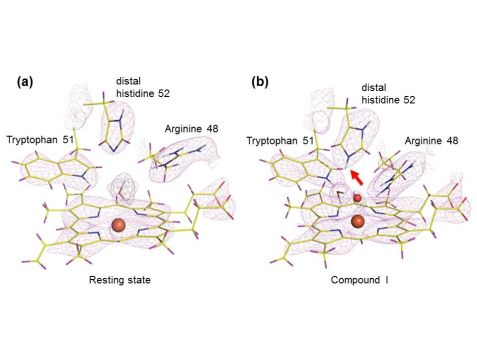
The structures showed that the distal histidine residue in the active site is neutral (single protonated) in the resting state but doubly protonated in Compound I, which was unexpected (see Fig. 3). The iron axial ligand in Compound I is an oxygen atom, and it is non-protonated. The oxygen forms hydrogen bonds to the residue Tryptophan 51 and Arginine 48. Our observations indicate that the widely assumed role of the distal histidine in Compound I formation needs to be reassessed and we proposed a possible alternative mechanisms for O-O bond cleavage [1].
This work shows the feasibility of using neutron cryo-crystallography for the clarification of the reaction mechanism in enzymatic pathways.
Reference:
[1] C. M. Casadei et al., Science 345, 193 (2014).
MLZ is a cooperation between:
 > Technische Universität München
> Technische Universität München > Helmholtz-Zentrum Hereon
> Helmholtz-Zentrum Hereon
 > Forschungszentrum Jülich
> Forschungszentrum Jülich
MLZ is a member of:
 > LENS
> LENS > ERF-AISBL
> ERF-AISBL
MLZ on social media: