MLZ is a cooperation between:
 > Technische Universität München
> Technische Universität München > Helmholtz-Zentrum Hereon
> Helmholtz-Zentrum Hereon
 > Forschungszentrum Jülich
> Forschungszentrum Jülich
MLZ is a member of:
 > LENS
> LENS > ERF-AISBL
> ERF-AISBL
MLZ on social media:

MLZ (eng)
Lichtenbergstr.1
85748 Garching
Illuminating the function of a biofuels enzyme
with neutrons
Q. Wan1, J. M. Parks2, B. L. Hanson3, S. Z. Fisher4, A. Ostermann5, T. E. Schrader6, D. E. Graham7,
L. Coates8, P. Langan8, A. Kovalevsky8
1Department of Physics, College of Science, Nanjing Agricultural University, Nanjing,China
2UT/ORNL Center for Molecular Biophysics, Biosciences Division, Oak Ridge National Laboratory, Oak Ridge, Tennesse, USA
3Chemistry Department, University of Toledo, Toledo, Ohio, USA
4Scientific Activities Division, European Spallation Source, Lund, Sweden
5Heinz Maier-Leibnitz Zentrum (MLZ), Technische Universität München, Garching, Germany
6Jülich Centre for Neutron Science (JCNS) at MLZ, Forschungszentrum Jülich GmbH, Garching, Germany
7Biosciences Division, Oak Ridge National Laboratory, Oak Ridge, Tennesse, USA
8Biology and Soft Matter Division, Oak Ridge National Laboratory, Oak Ridge, Tennesse, USA
Plants can be converted into a variety of renewable products such as fuels. To efficiently deconstruct biomass material, efficient enzymes such as glycoside hydrolases are being investigated. Glycoside hydrolases apply acid/base chemistry to catalyze the decomposition of complex carbohydrates by accepting protons from the solvent and donating them to substrates, cellulose and hemicellulose. However, it is not known how the catalytic acid residue acquires a proton and efficiently transfers it to the substrate. We used macromolecular neutron crystallography to directly determine protonation states of the active site residues of the enzyme xylanase at multiple pH values. We characterized the initial stage of the glycoside hydrolysis, in which the catalytic glutamate residue obtains a proton from water, and then delivers it to the glycosidic oxygen. The catalytic glutamate cycles between two conformations, “upward” and “downward”, with the energy barrier of ~4 kcal/mol, but is protonated only in the “downward” orientation with higher pKa. These findings shed light on xylanase function and will aid in protein engineering efforts.
Protons are key players in glycoside hydrolysis
Catalysis by biomacromolecules called enzymes frequently involves proton transfer. To fully understand enzyme chemistry, accurate hydrogen atom positions must be mapped. One class of enzymes are glycoside hydrolases (GHs) that cleave, or hydrolyze, the glycosidic bond connecting two sugar units in a polysaccharide. The glycosidic bond hydrolysis by GHs is accomplished with rate enhancements of about 1018, placing them among the most efficient biocatalysts in Nature. GHs are utilized in the production of biofuels from plant biomass that requires decomposition of natural polysaccharides into fermentable sugars [1]. Understanding their catalytic mechanisms and ligand binding is of paramount importance for enzyme design to improve biofuel production.
GH enzymes enhance the rate of glycoside hydrolysis by using a general acid, usually the carboxylic acid of a glutamate (Glu) residue, which donates a proton to the leaving group oxygen. A second Glu residue located nearby acts as a nucleophile to attack the carbon atom of the glycosidic bond and generate a covalent intermediate. The next reaction step involves the general acid Glu switching its role to act as a general base, assisting an incoming water molecule to break the covalent intermediate. In the reactant state, the general acid must be weaker than the nucleophilic Glu, implying that the former has to be protonated, with a pKa greater than 6, and the nucleophile must be deprotonated, with a pKa less than 5. However, the protonation states of amino acid residues, as well as the distribution of hydrogen bonds, have not been directly observed in GH enzymes, and the changes in protonation predicted by the reaction mechanism have only been inferred from indirect measurements.
Atomic-level picture of a GH enzyme by neutron crystallography
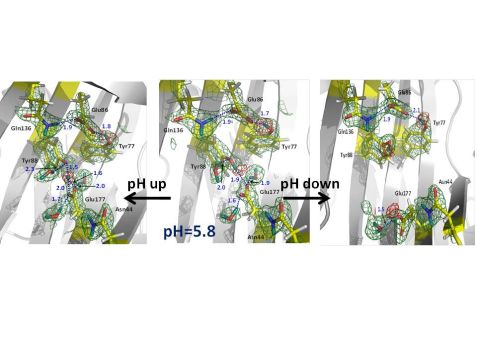
Figure 1: Nuclear scattering length density maps (green mesh) of the enzyme active site. The red mesh indicates the location of deuterium atoms, showing protonation (deuteration) of the catalytic glutamate Glu177 only in the downward conformation. Atomic distances are given in Å (blue).
Hydrogen atoms are crucial players in the mechanism of glycoside hydrolysis, participating in directional hydrogen bonding and proton transfer events. However, hydrogen atoms are virtually invisible to X-rays due to their poor scattering power. Neutrons represent a superb probe to locate hydrogen atoms and obtain atomic details of their movements in biomacromolecules. Locating hydrogen (deuterium (D)) atoms in macromolecular neutron structures is straightforward even at medium resolutions with a dmin of 2.0 – 2.5 Å. Thus, macromolecular neutron crystallography has been used to better understand the catalytic mechanism of glycoside hydrolysis. The research team obtained five neutron structures of a family 11 GH, xylanase, in the ligand-free and ligand-bound states to find out how the general acid Glu (Glu177) receives a proton from water and then delivers it to the substrate [2]. Surprisingly, the neutron structure at pD 6.2 (pD = pH + 0.4), in which the Glu177 side chain is in the “upward” conformation facing the nucleophilic Glu86, indicates that it is not protonated, even though its pKa is 7 (Fig. 1, middle). Remarkably, no protonation of Glu177 is observed in Asn44Asp mutant, although a general acid pKa of 8 is expected. Interestingly, at higher pD of 8.9, Glu177 is engaged in a strong hydrogen bond with a nearby tyrosine residue Tyr88 so that the phenolic D atom is found midway between oxygen atoms (Fig. 1, left side), indicating the hydrogen bond would also be strong at pD 6 and making Glu177 a stronger rather than a weaker acid. As it turns out, Glu177 side chain can have another conformation, rotated “downward” into a hydrophobic pocket. pKa calculations estimate that Glu177 becomes a weaker acid, with higher pKa, when it rotates “downward”. The energy barrier required (4.3 kcal/mol) is low so that the “downward” and “upward” conformations are in dynamic equilibrium. To locate the hydrogen on Glu177 in the “downward” conformation, a neutron structure at pD of 4.8 was obtained. Indeed, Glu177 was observed in the neutral, protonated (deuterated) form (Fig. 1, right side).
Glutamic acid conformational dynamics is essential for GH function
Based on the neutron structures, pKa calculations and molecular dynamics simulations it is proposed that Glu177 obtains a proton from water when in the “downward” conformation. The general acid Glu177 side chain dynamically moves between the “downward” conformation, with high pKa facilitating its protonation, and the “upward” conformation, with low pKa facilitating the delivery of a proton to the glycosidic oxygen. These findings possibly resolve decades of confusion surrounding the question of why the general acid and nucleophilic glutamates in family 11 GH enzymes, while having similar hydrogen bonding interactions and solvent accessibilities, exhibit very different pKa values and assume their corresponding catalytic roles. A better understanding of the GH catalytic mechanism provided by this neutron crystallographic study in combination with theoretical calculations will improve protein engineering efforts to design more efficient GH enzymes.
References:
[1] D. B. Wilson, Curr. Opin. Biotechnol. 20, 295 (2009).
[2] Q. Wan et al., Proc. Natl. Acad. Sci. U.S.A. 112, 12384 (2015).
MLZ is a cooperation between:
 > Technische Universität München
> Technische Universität München > Helmholtz-Zentrum Hereon
> Helmholtz-Zentrum Hereon
 > Forschungszentrum Jülich
> Forschungszentrum Jülich
MLZ is a member of:
 > LENS
> LENS > ERF-AISBL
> ERF-AISBL
MLZ on social media:


